Gold »
PDB 1a52-3k3g »
1un1 »
Gold in PDB 1un1: Xyloglucan Endotransglycosylase Native Structure.
Enzymatic activity of Xyloglucan Endotransglycosylase Native Structure.
All present enzymatic activity of Xyloglucan Endotransglycosylase Native Structure.:
2.4.1.207;
2.4.1.207;
Protein crystallography data
The structure of Xyloglucan Endotransglycosylase Native Structure., PDB code: 1un1
was solved by
P.Johansson,
H.Brumer,
A.Kallas,
H.Henriksson,
S.Denman,
T.T.Teeri,
T.A.Jones,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 24.77 / 2.1 |
| Space group | P 63 |
| Cell size a, b, c (Å), α, β, γ (°) | 188.762, 188.762, 45.973, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 20.6 / 23.2 |
Gold Binding Sites:
The binding sites of Gold atom in the Xyloglucan Endotransglycosylase Native Structure.
(pdb code 1un1). This binding sites where shown within
5.0 Angstroms radius around Gold atom.
In total only one binding site of Gold was determined in the Xyloglucan Endotransglycosylase Native Structure., PDB code: 1un1:
In total only one binding site of Gold was determined in the Xyloglucan Endotransglycosylase Native Structure., PDB code: 1un1:
Gold binding site 1 out of 1 in 1un1
Go back to
Gold binding site 1 out
of 1 in the Xyloglucan Endotransglycosylase Native Structure.
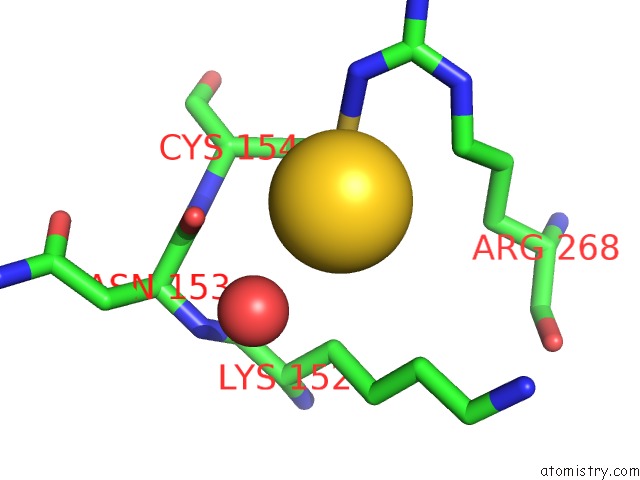
Mono view
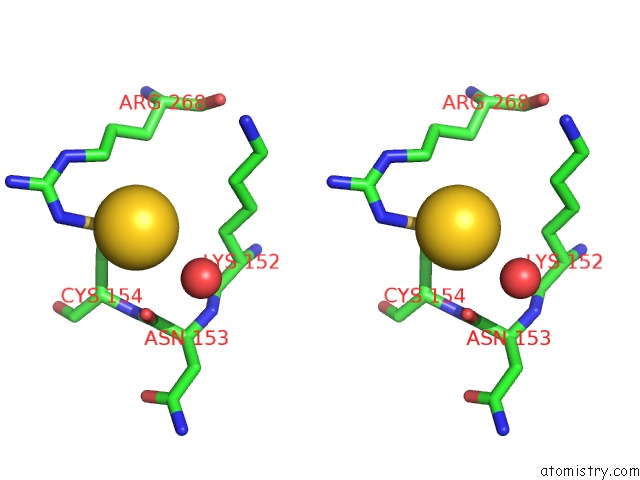
Stereo pair view

Mono view

Stereo pair view
A full contact list of Gold with other atoms in the Au binding
site number 1 of Xyloglucan Endotransglycosylase Native Structure. within 5.0Å range:
|
Reference:
P.Johansson,
H.Brumer,
M.Baumann,
A.Kallas,
H.Henriksson,
S.Denman,
T.T.Teeri,
T.A.Jones.
Crystal Structures of A Poplar Xyloglucan Endotransglycosylase Reveal Details of Transglycosylation Acceptor Binding Plant Cell V. 16 874 2004.
ISSN: ISSN 1040-4651
PubMed: 15020748
DOI: 10.1105/TPC.020065
Page generated: Mon Jul 7 01:19:08 2025
ISSN: ISSN 1040-4651
PubMed: 15020748
DOI: 10.1105/TPC.020065
Last articles
Cl in 5FKMCl in 5FJV
Cl in 5FKL
Cl in 5FJK
Cl in 5FJL
Cl in 5FJJ
Cl in 5FJG
Cl in 5FHO
Cl in 5FJH
Cl in 5FJ3